Life Sciences Use Case: HGST Active Archive and Panasas ActiveStor
The iRODS Consortium is pleased to share a combined life sciences use case from our members - HGST and Panasas.
Accelerating research requires fast processes and fast disk. iRODS, atop HGST Active Archive and Panasas ActiveStor, provides a uniform namespace and common policy engine to manage the demanding tiering required for modern life sciences research.
Life Sciences / HPC Storage and Data Management Architecture
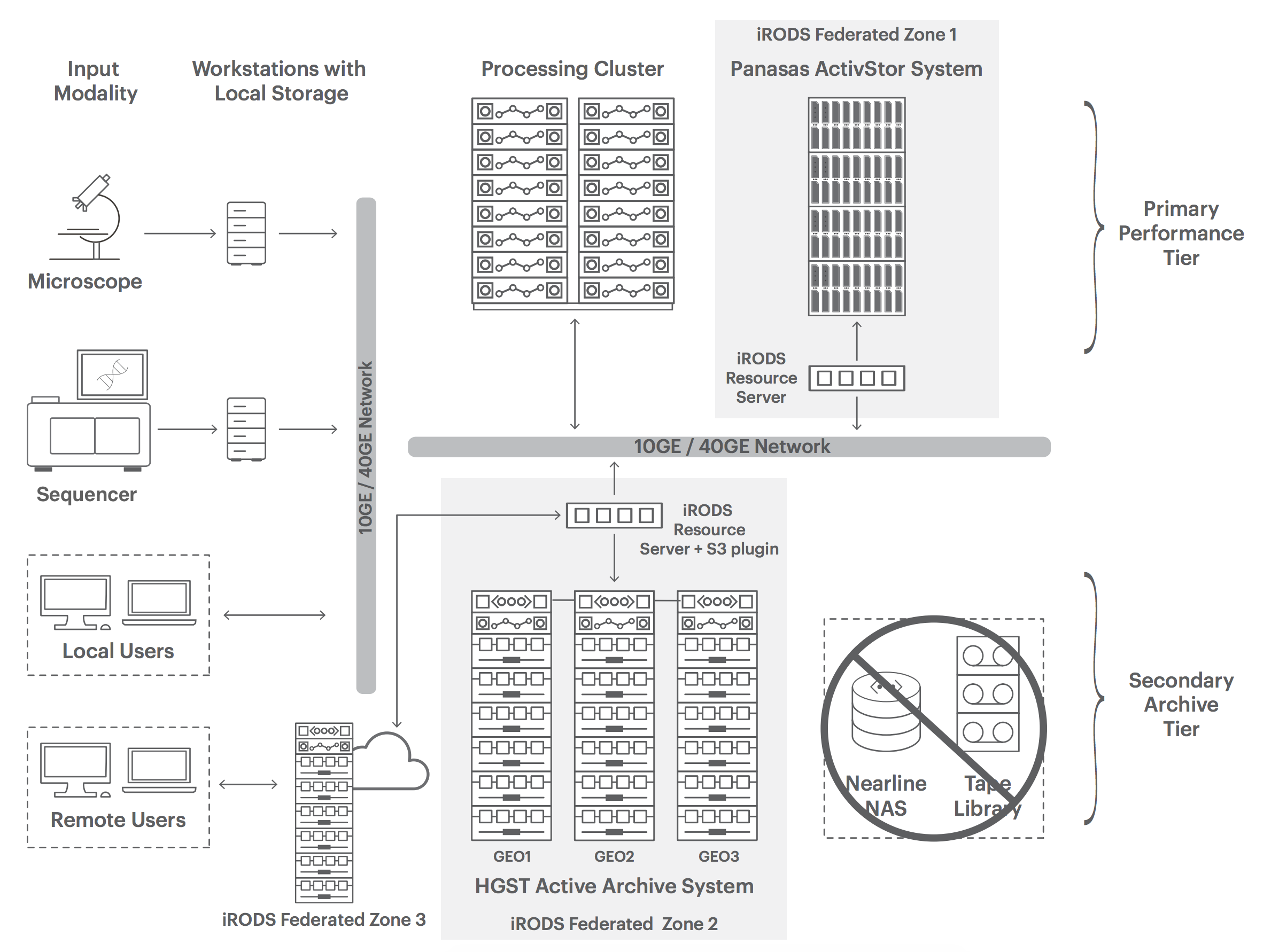
Speed Research Discovery with Comprehensive Storage and Data Management
The rate of progress in life sciences research is accelerating exponentially leading to important advances in healthcare, agriculture, climate science and more, but those advances also create a mountain of data. IT managers supporting these efforts are being challenged to provide researchers the right solution that will accelerate their work. Complex simulations can take days or weeks to run. When simulations take longer, discoveries are delayed slowing analysis and ultimately the commercialization of the findings. Faster simulations allow more complex models to be run – leading to improved discoveries through bioinformatics. A key to accelerating life science application performance is implementing a high performance computing (HPC) infrastructure that eliminates computing and storage bottlenecks, enables better collaboration, and preserves simplicity so researchers focus their efforts on discovery.
Conventional Storage is Slowing the Progress
Storage performance and data management are major causes of computing bottlenecks. The volume and type of data generated by modern lab equipment along with varying application and workflow requirements, makes implementing the right solution all the more challenging. In some cases, data is generated in one place and kept there, while other times the data is generated by many researchers around the world whose results and expertise must be pooled together to achieve the biggest benefit. Furthermore, HPC environments place special demands on storage with compute clusters that can have hundreds of nodes and thousands of cores working in parallel. Technical computing applications tend to be I/O bound with large numbers of sequential and random rapid read/write operations that can exhaust conventional storage, resulting in workflow bottlenecks, costly islands of storage, increased management effort, and longer time-to-discovery. A better approach is needed.
Eliminate Storage Performance Bottlenecks with Parallel Access
Panasas has an advanced performance scale-out NAS solution called ActiveStor that is designed to maximize mixed workload performance in HPC environments. Based on a fifth-generation architecture and parallel file system, application clients have simultaneous fast direct parallel access to large and growing datasets, avoiding the need to copy datasets locally to the compute cluster prior to processing. Direct parallel data access is also important because in the case of genomics, while sequencers often generate data in single streams, analysis of sequencer data can be done in parallel with many clients reading and writing directly to storage. In addition, up to 90% of metadata-related operations happen outside the data path, minimizing data access impact, resulting in faster workflows.
Object Storage for Massive Data Growth and Global Collaboration
Built using next generation object storage technology, the HGST Active Archive System enables research organizations to help cost-effectively manage enormous data growth. Serving as the capacity optimized secondary archive tier (Figure 1), the system’s industry leading durability and data integrity makes it ideal for long-term data preservation. The system is simple to deploy and manage and can be easily scaled over time. IT management overhead is minimized with automated self-healing and proactive data integrity checks. Deployed in 3-site-geospread configuration, data is efficiently spread across three sites making it ideal for collaboration across distributed researchers. Integration with HPC workflows is easy using iRODS and its S3 resource server plugin.
Data Management at Scale in a Distributed Environment
Data-intensive technical computing applications such as in life sciences, require efficient, secure and cost effective data management in a wide-area distributed environment. Datasets are often shared by a global community of researchers that need to easily find and transfer subsets of data to a local or remote resource such as a private or public cloud for further processing. Open source data management software called iRODS (Integrated Rule-Oriented Data System) is increasingly used in a variety of technical computing workflows.
iRODS virtualizes data storage resources by putting a unified namespace on files regardless of where they are located, making them appear to the user as one project. Data discovery is made easy using a metadata catalog that describes every file, every directory, and every storage resource in the iRODS data grid, including federated grids (Figure 1). Data workflows can be automated with a rules engine that permits any action to be initiated by any trigger on any server or client in the grid. Collaboration is made secure and easy with configurable user authentication and SSL, users need only to be logged in to their home grid to access data on a remote grid. iRODS also helps control costs by allowing data to be aligned to the right storage technology through rules-based tiering. For instance, the majority of data can be stored on a capacity optimized object storage active archive tier and automatically moved to the high-performance scale-out file tier – significantly reducing CapEx.
Terrell Russell